Download RIRA
GetRira.RmdThis vignette demonstrates how to download the RIRA Rhesus Macaque scRNA-seq Atlas as a Seurat object.
Download RIRA Data:
This example downloads the processed Seurat object from GEO. This is a large file (~10GB); however, it contains pre-computed dimensionality reductions and metadata. It is also possible to download the count matrix from GEO, such as with the GEOquery R package. In addition to the entire Seurat object, just the metadata (~67mb) is available to download as a table. This table could be merged with the raw counts if you only are interested in specific information.
Due to the large size, in practice it might be more useful to download the ‘T_NK’, ‘Bcell’, or ‘Myeloid’ subsets.
seuratObj <- DownloadFromGEO(subset = 'All', outfile = 'RIRA_All.rds')
# or:
seuratObj <- DownloadFromGEO(subset = 'T_NK', outfile = 'RIRA_All.rds')Basic Exploration
As a seurat object, the RIRA data is compatible with a wide range of scRNA-seq analysis and visualization tools. Below are examples, which take advantage of the metadata and cell classifications included with this dataset:
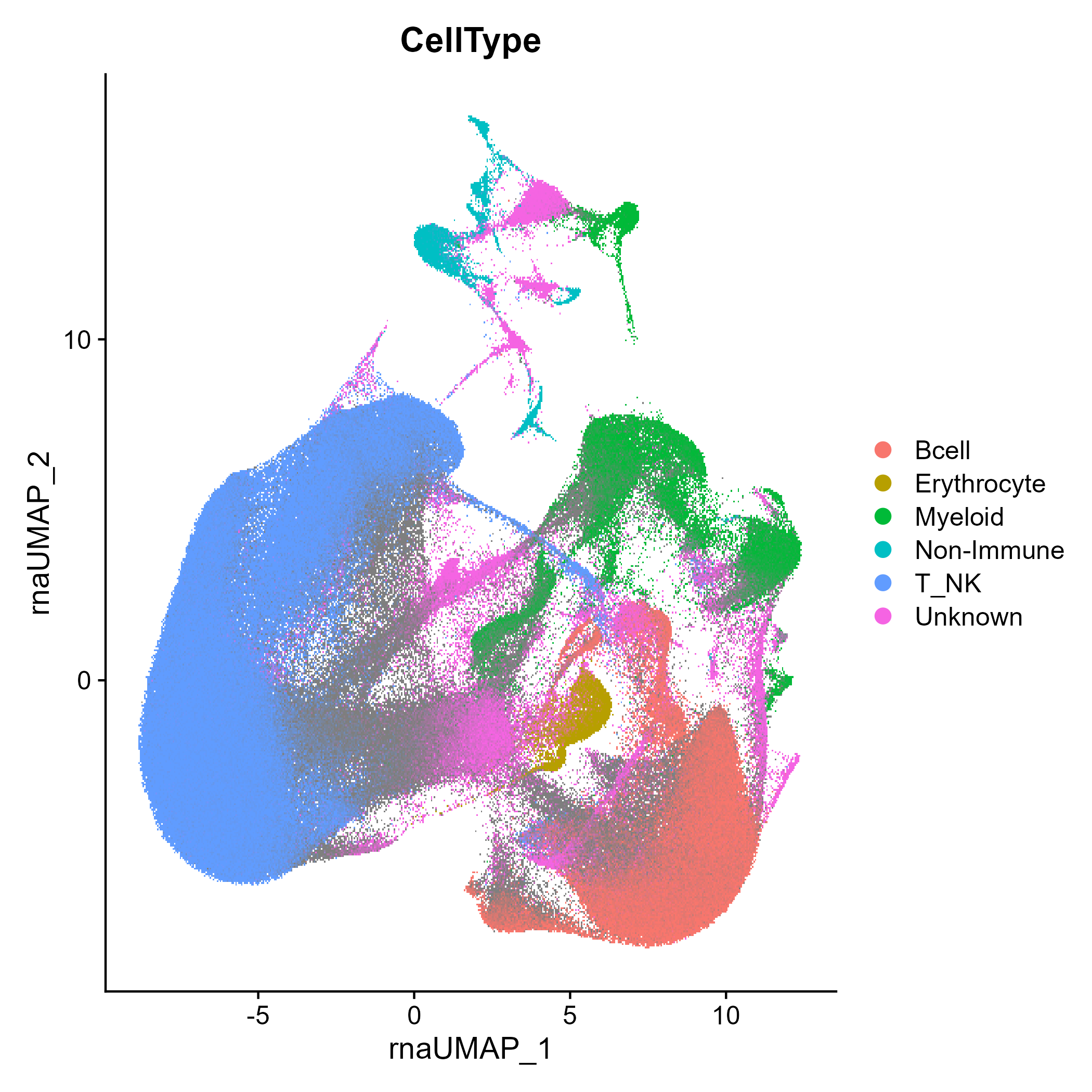
This is a coarse cell type classification (see the
vignette('CellType') for more information on how this is
calculated and could be used on other data):
DimPlot(seuratObj, group.by = 'RIRA_Immune_v2.cellclass')